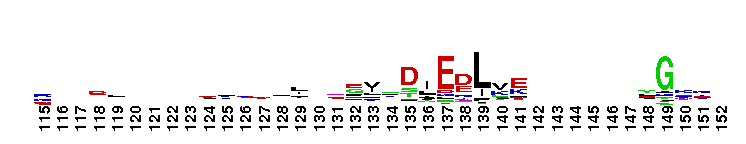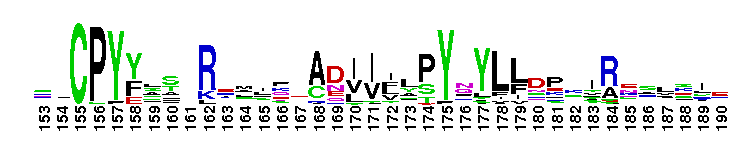| ||||||||||||||||||||||||
Tips:  Range on the Protein: Protein ID Protein Position Domain Position:  No Conserved Features/Sites Found for DEAD_2 No Conserved Features/Sites Found for DEAD_2
|
|---|
Weblogos are Copyright (c) 2002 Regents of the University of California
| DMDM_info@umbc.edu | 1000 Hilltop Circle, Baltimore, MD 21250 | Department of Biological Sciences | Phone: 410-455-2258 |




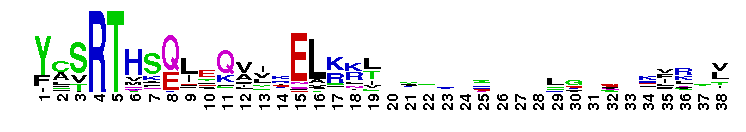
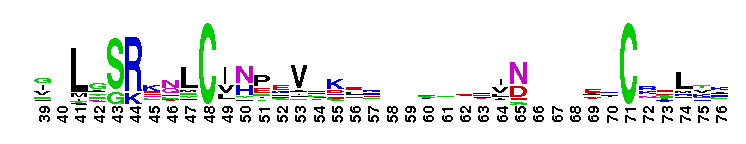
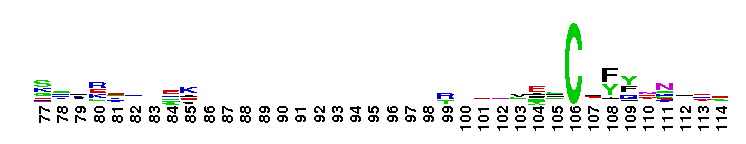
 DEAD_2. This represents a conserved region within a number of RAD3-like DNA-binding helicases that are seemingly ubiquitous - members include proteins of eukaryotic, bacterial and archaeal origin. RAD3 is involved in nucleotide excision repair, and forms part of the transcription factor TFIIH in yeast.
DEAD_2. This represents a conserved region within a number of RAD3-like DNA-binding helicases that are seemingly ubiquitous - members include proteins of eukaryotic, bacterial and archaeal origin. RAD3 is involved in nucleotide excision repair, and forms part of the transcription factor TFIIH in yeast. No pairwise interactions found for the domain DEAD_2
No pairwise interactions found for the domain DEAD_2