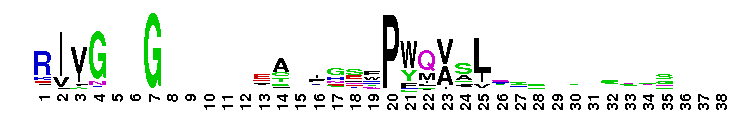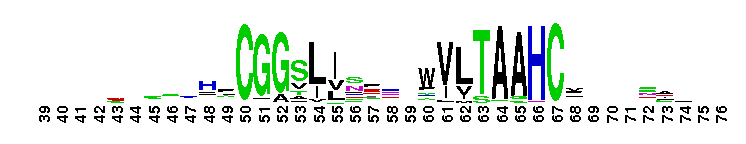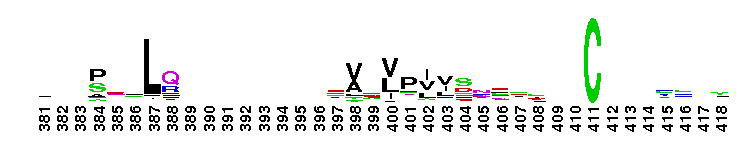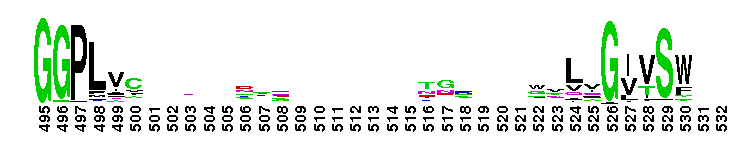| |||||||||||||||||||||||||||||||||||||||||||||||||||||
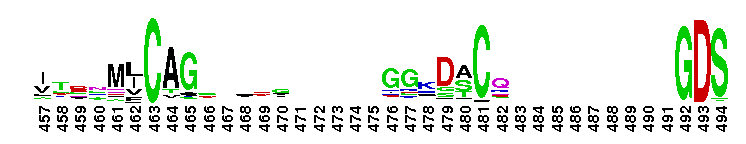
Tips:  Range on the Protein: Protein ID Protein Position Domain Position:  No Conserved Features/Sites Found for Tryp_SPc No Conserved Features/Sites Found for Tryp_SPc
|
|---|
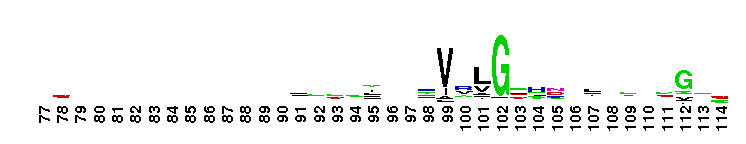
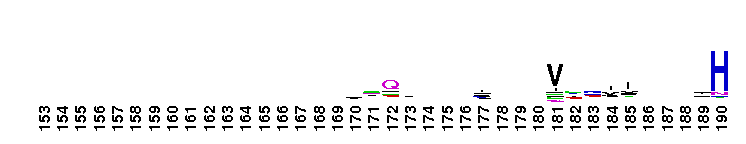
Weblogos are Copyright (c) 2002 Regents of the University of California
| DMDM_info@umbc.edu | 1000 Hilltop Circle, Baltimore, MD 21250 | Department of Biological Sciences | Phone: 410-455-2258 |




 Trypsin-like serine protease. Many of these are synthesised as inactive precursor zymogens that are cleaved during limited proteolysis to generate their active forms. A few, however, are active as single chain molecules, and others are inactive due to substitutions of the catalytic triad residues.
Trypsin-like serine protease. Many of these are synthesised as inactive precursor zymogens that are cleaved during limited proteolysis to generate their active forms. A few, however, are active as single chain molecules, and others are inactive due to substitutions of the catalytic triad residues. No pairwise interactions found for the domain Tryp_SPc
No pairwise interactions found for the domain Tryp_SPc