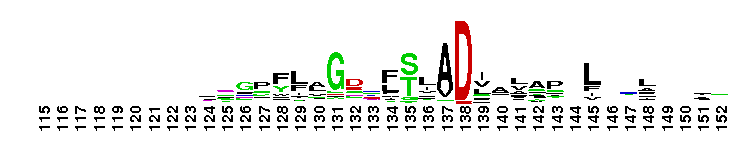| ||||||||||||||||||||||||||||
Tips:  Range on the Protein: Protein ID Protein Position Domain Position: 
|
|---|
Weblogos are Copyright (c) 2002 Regents of the University of California
| DMDM_info@umbc.edu | 1000 Hilltop Circle, Baltimore, MD 21250 | Department of Biological Sciences | Phone: 410-455-2258 |




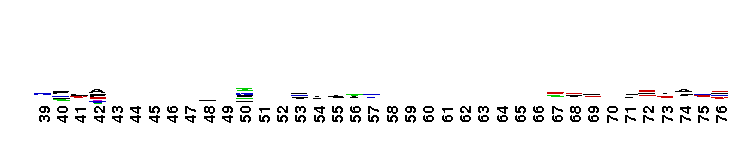
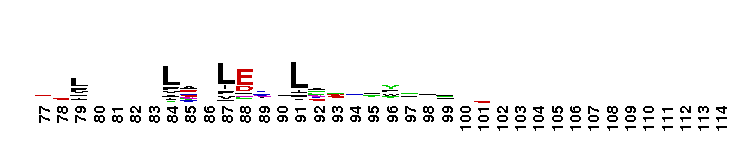
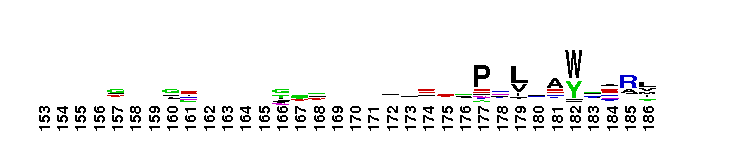
 C-terminal, alpha helical domain of the Glutathione S-transferase family. Glutathione S-transferase (GST) family, C-terminal alpha helical domain; a large, diverse group of cytosolic dimeric proteins involved in cellular detoxification by catalyzing the conjugation of glutathione (GSH) with a wide range of endogenous and xenobiotic alkylating agents, including carcinogens, therapeutic drugs, environmental toxins and products of oxidative stress. In addition, GSTs also show GSH peroxidase activity and are involved in the synthesis of prostaglandins and leukotrienes. This family, also referred to as soluble GSTs, is the largest family of GSH transferases and is only distantly related to the mitochondrial GSTs (GSTK). Soluble GSTs bear no structural similarity to microsomal GSTs (MAPEG family) and display additional activities unique to their group, such as catalyzing thiolysis, reduction and isomerization of certain compounds. The GST fold contains an N-terminal thioredoxin-fold domain and a C-terminal alpha helical domain, with an active site located in a cleft between the two domains. GSH binds to the N-terminal domain while the hydrophobic substrate occupies a pocket in the C-terminal domain. Based on sequence similarity, different classes of GSTs have been identified, which display varying tissue distribution, substrate specificities and additional specific activities. In humans, GSTs display polymorphisms which may influence individual susceptibility to diseases such as cancer, arthritis, allergy and sclerosis. Some GST family members with non-GST functions include glutaredoxin 2, the CLIC subfamily of anion channels, prion protein Ure2p, crystallins, metaxins, stringent starvation protein A, and aminoacyl-tRNA synthetases.
C-terminal, alpha helical domain of the Glutathione S-transferase family. Glutathione S-transferase (GST) family, C-terminal alpha helical domain; a large, diverse group of cytosolic dimeric proteins involved in cellular detoxification by catalyzing the conjugation of glutathione (GSH) with a wide range of endogenous and xenobiotic alkylating agents, including carcinogens, therapeutic drugs, environmental toxins and products of oxidative stress. In addition, GSTs also show GSH peroxidase activity and are involved in the synthesis of prostaglandins and leukotrienes. This family, also referred to as soluble GSTs, is the largest family of GSH transferases and is only distantly related to the mitochondrial GSTs (GSTK). Soluble GSTs bear no structural similarity to microsomal GSTs (MAPEG family) and display additional activities unique to their group, such as catalyzing thiolysis, reduction and isomerization of certain compounds. The GST fold contains an N-terminal thioredoxin-fold domain and a C-terminal alpha helical domain, with an active site located in a cleft between the two domains. GSH binds to the N-terminal domain while the hydrophobic substrate occupies a pocket in the C-terminal domain. Based on sequence similarity, different classes of GSTs have been identified, which display varying tissue distribution, substrate specificities and additional specific activities. In humans, GSTs display polymorphisms which may influence individual susceptibility to diseases such as cancer, arthritis, allergy and sclerosis. Some GST family members with non-GST functions include glutaredoxin 2, the CLIC subfamily of anion channels, prion protein Ure2p, crystallins, metaxins, stringent starvation protein A, and aminoacyl-tRNA synthetases. No pairwise interactions are available for this conserved domain.
No pairwise interactions are available for this conserved domain.