| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
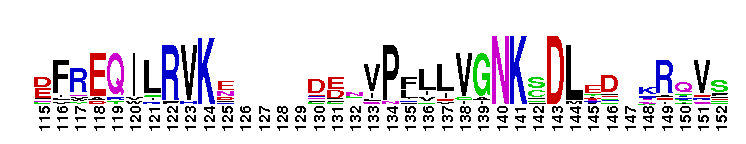
Tips:  Range on the Protein: Protein ID Protein Position Domain Position: 
|
|---|
Weblogos are Copyright (c) 2002 Regents of the University of California
| DMDM_info@umbc.edu | 1000 Hilltop Circle, Baltimore, MD 21250 | Department of Biological Sciences | Phone: 410-455-2258 |




 Ral (Ras-like) family containing highly homologous RalA and RalB. The Ral (Ras-like) subfamily consists of the highly homologous RalA and RalB. Ral proteins are believed to play a crucial role in tumorigenesis, metastasis, endocytosis, and actin cytoskeleton dynamics. Despite their high sequence similarity (>80% sequence identity), nonoverlapping and opposing functions have been assigned to RalA and RalBs in tumor migration. In human bladder and prostate cancer cells, RalB promotes migration while RalA inhibits it. A Ral-specific set of GEFs has been identified that are activated by Ras binding. This RalGEF activity is enhanced by Ras binding to another of its target proteins, phosphatidylinositol 3-kinase (PI3K). Ral effectors include RLIP76/RalBP1, a Rac/cdc42 GAP, and the exocyst (Sec6/8) complex, a heterooctomeric protein complex that is involved in tethering vesicles to specific sites on the plasma membrane prior to exocytosis. In rat kidney cells, RalB is required for functional assembly of the exocyst and for localizing the exocyst to the leading edge of migrating cells. In human cancer cells, RalA is required to support anchorage-independent proliferation and RalB is required to suppress apoptosis. RalA has been shown to localize to the plasma membrane while RalB is localized to the intracellular vesicles. Most Ras proteins contain a lipid modification site at the C-terminus, with a typical sequence motif CaaX, where a = an aliphatic amino acid and X = any amino acid. Lipid binding is essential for membrane attachment, a key feature of most Ras proteins. Due to the presence of truncated sequences in this CD, the lipid modification site is not available for annotation.
Ral (Ras-like) family containing highly homologous RalA and RalB. The Ral (Ras-like) subfamily consists of the highly homologous RalA and RalB. Ral proteins are believed to play a crucial role in tumorigenesis, metastasis, endocytosis, and actin cytoskeleton dynamics. Despite their high sequence similarity (>80% sequence identity), nonoverlapping and opposing functions have been assigned to RalA and RalBs in tumor migration. In human bladder and prostate cancer cells, RalB promotes migration while RalA inhibits it. A Ral-specific set of GEFs has been identified that are activated by Ras binding. This RalGEF activity is enhanced by Ras binding to another of its target proteins, phosphatidylinositol 3-kinase (PI3K). Ral effectors include RLIP76/RalBP1, a Rac/cdc42 GAP, and the exocyst (Sec6/8) complex, a heterooctomeric protein complex that is involved in tethering vesicles to specific sites on the plasma membrane prior to exocytosis. In rat kidney cells, RalB is required for functional assembly of the exocyst and for localizing the exocyst to the leading edge of migrating cells. In human cancer cells, RalA is required to support anchorage-independent proliferation and RalB is required to suppress apoptosis. RalA has been shown to localize to the plasma membrane while RalB is localized to the intracellular vesicles. Most Ras proteins contain a lipid modification site at the C-terminus, with a typical sequence motif CaaX, where a = an aliphatic amino acid and X = any amino acid. Lipid binding is essential for membrane attachment, a key feature of most Ras proteins. Due to the presence of truncated sequences in this CD, the lipid modification site is not available for annotation. No pairwise interactions are available for this conserved domain.
No pairwise interactions are available for this conserved domain.