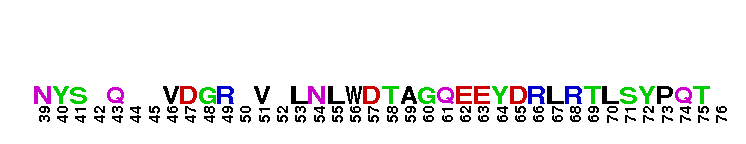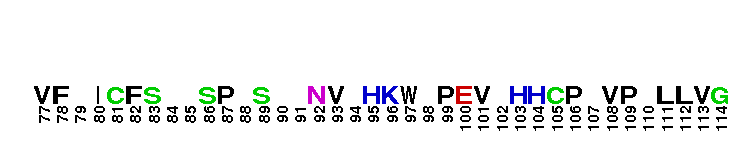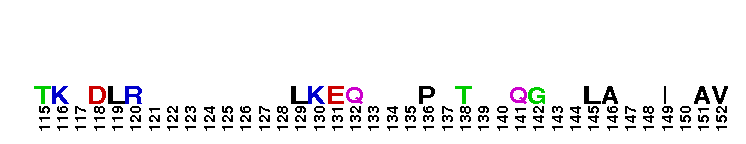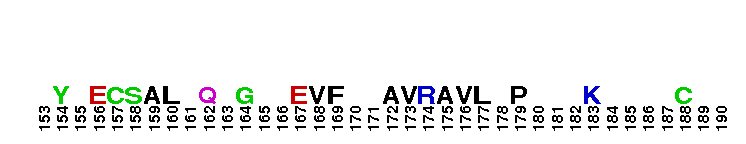| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tips:  Range on the Protein: Protein ID Protein Position Domain Position: 
|
|---|
Weblogos are Copyright (c) 2002 Regents of the University of California
| DMDM_info@umbc.edu | 1000 Hilltop Circle, Baltimore, MD 21250 | Department of Biological Sciences | Phone: 410-455-2258 |




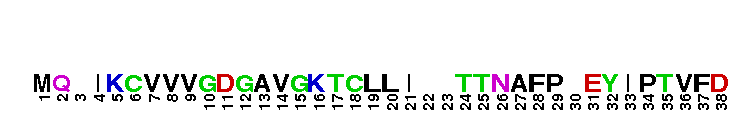
 Ras homolog family, member G (RhoG) of small guanosine triphosphatases (GTPases). RhoG is a GTPase with high sequence similarity to members of the Rac subfamily, including the regions involved in effector recognition and binding. However, RhoG does not bind to known Rac1 and Cdc42 effectors, including proteins containing a Cdc42/Rac interacting binding (CRIB) motif. Instead, RhoG interacts directly with Elmo, an upstream regulator of Rac1, in a GTP-dependent manner and forms a ternary complex with Dock180 to induce activation of Rac1. The RhoG-Elmo-Dock180 pathway is required for activation of Rac1 and cell spreading mediated by integrin, as well as for neurite outgrowth induced by nerve growth factor. Thus RhoG activates Rac1 through Elmo and Dock180 to control cell morphology. RhoG has also been shown to play a role in caveolar trafficking and has a novel role in signaling the neutrophil respiratory burst stimulated by G protein-coupled receptor (GPCR) agonists. Most Rho proteins contain a lipid modification site at the C-terminus, with a typical sequence motif CaaX, where a = an aliphatic amino acid and X = any amino acid. Lipid binding is essential for membrane attachment, a key feature of most Rho proteins.
Ras homolog family, member G (RhoG) of small guanosine triphosphatases (GTPases). RhoG is a GTPase with high sequence similarity to members of the Rac subfamily, including the regions involved in effector recognition and binding. However, RhoG does not bind to known Rac1 and Cdc42 effectors, including proteins containing a Cdc42/Rac interacting binding (CRIB) motif. Instead, RhoG interacts directly with Elmo, an upstream regulator of Rac1, in a GTP-dependent manner and forms a ternary complex with Dock180 to induce activation of Rac1. The RhoG-Elmo-Dock180 pathway is required for activation of Rac1 and cell spreading mediated by integrin, as well as for neurite outgrowth induced by nerve growth factor. Thus RhoG activates Rac1 through Elmo and Dock180 to control cell morphology. RhoG has also been shown to play a role in caveolar trafficking and has a novel role in signaling the neutrophil respiratory burst stimulated by G protein-coupled receptor (GPCR) agonists. Most Rho proteins contain a lipid modification site at the C-terminus, with a typical sequence motif CaaX, where a = an aliphatic amino acid and X = any amino acid. Lipid binding is essential for membrane attachment, a key feature of most Rho proteins. No pairwise interactions are available for this conserved domain.
No pairwise interactions are available for this conserved domain.