| |||||||||||||||||||
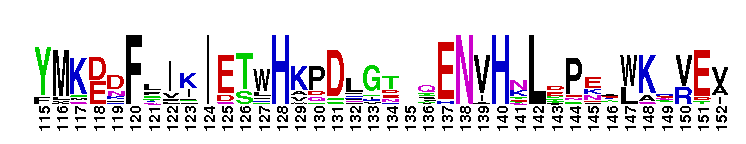
Tips:  Range on the Protein: Protein ID Protein Position Domain Position: 
|
|---|
Weblogos are Copyright (c) 2002 Regents of the University of California
| DMDM_info@umbc.edu | 1000 Hilltop Circle, Baltimore, MD 21250 | Department of Biological Sciences | Phone: 410-455-2258 |




 Lipid-binding SRPBCC domain of mammalian PITPNA, -B, and related proteins (Class I PITPs). This subgroup includes the SRPBCC (START/RHO_alpha_C/PITP/Bet_v1/CoxG/CalC) domain of mammalian Class 1 phosphatidylinositol transfer proteins (PITPs), PITPNA/PITPalpha and PITPNB/PITPbeta, Drosophila vibrator, and related proteins. These are single domain proteins belonging to the PITP family of lipid transfer proteins, and to the SRPBCC domain superfamily of proteins that bind hydrophobic ligands. SRPBCC domains have a deep hydrophobic ligand-binding pocket. In vitro, PITPs bind phosphatidylinositol (PtdIns), as well as phosphatidylcholine (PtdCho) but with a lower affinity. They transfer these lipids from one membrane compartment to another. The cellular roles of PITPs include inositol lipid signaling, PtdIns metabolism, and membrane trafficking. In addition, PITPNB transfers sphingomyelin in vitro, with a low affinity. PITPNA is found chiefly in the nucleus and cytoplasm; it is enriched in the brain and predominantly localized in the axons. A reduced expression of PITPNA contributes to the neurodegenerative phenotype of the mouse vibrator mutation. The role of PITPNA in vivo may be to provide PtdIns for localized PI3K-dependent signaling, thereby controlling the polarized extension of axonal processes. PITPNA homozygous null mice die soon after birth from complicated organ failure, including intestinal and hepatic steatosis, hypoglycemia, and spinocerebellar disease. PITPNB is associated with the Golgi and ER, and is highly expressed in the liver. Deletion of the PITPNB gene results in embryonic lethality. The PtdIns and PtdCho exchange activity of PITPNB is required for COPI-mediated retrograde transport from the Golgi to the ER. Drosophila vibrator localizes to the ER, and has an essential role in cytokinesis during mitosis and meiosis.
Lipid-binding SRPBCC domain of mammalian PITPNA, -B, and related proteins (Class I PITPs). This subgroup includes the SRPBCC (START/RHO_alpha_C/PITP/Bet_v1/CoxG/CalC) domain of mammalian Class 1 phosphatidylinositol transfer proteins (PITPs), PITPNA/PITPalpha and PITPNB/PITPbeta, Drosophila vibrator, and related proteins. These are single domain proteins belonging to the PITP family of lipid transfer proteins, and to the SRPBCC domain superfamily of proteins that bind hydrophobic ligands. SRPBCC domains have a deep hydrophobic ligand-binding pocket. In vitro, PITPs bind phosphatidylinositol (PtdIns), as well as phosphatidylcholine (PtdCho) but with a lower affinity. They transfer these lipids from one membrane compartment to another. The cellular roles of PITPs include inositol lipid signaling, PtdIns metabolism, and membrane trafficking. In addition, PITPNB transfers sphingomyelin in vitro, with a low affinity. PITPNA is found chiefly in the nucleus and cytoplasm; it is enriched in the brain and predominantly localized in the axons. A reduced expression of PITPNA contributes to the neurodegenerative phenotype of the mouse vibrator mutation. The role of PITPNA in vivo may be to provide PtdIns for localized PI3K-dependent signaling, thereby controlling the polarized extension of axonal processes. PITPNA homozygous null mice die soon after birth from complicated organ failure, including intestinal and hepatic steatosis, hypoglycemia, and spinocerebellar disease. PITPNB is associated with the Golgi and ER, and is highly expressed in the liver. Deletion of the PITPNB gene results in embryonic lethality. The PtdIns and PtdCho exchange activity of PITPNB is required for COPI-mediated retrograde transport from the Golgi to the ER. Drosophila vibrator localizes to the ER, and has an essential role in cytokinesis during mitosis and meiosis. No pairwise interactions are available for this conserved domain.
No pairwise interactions are available for this conserved domain.