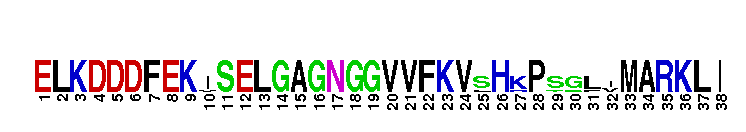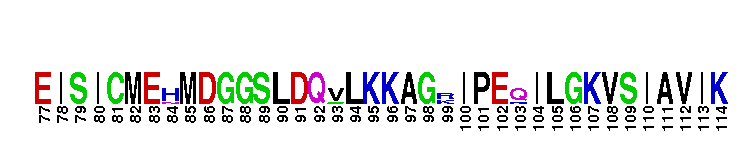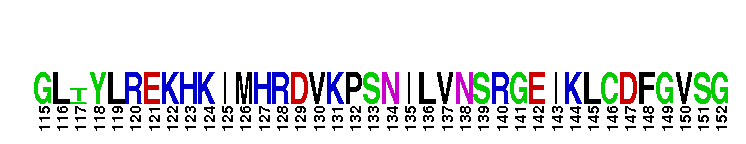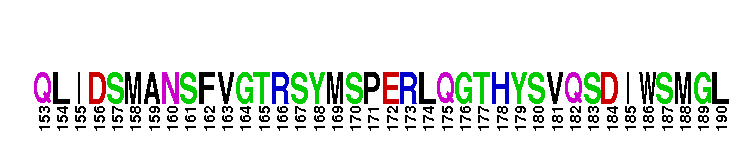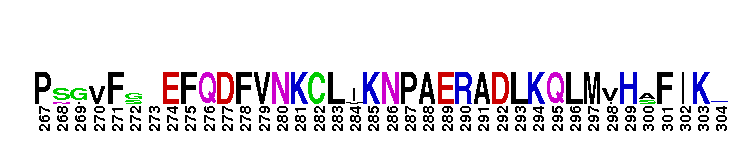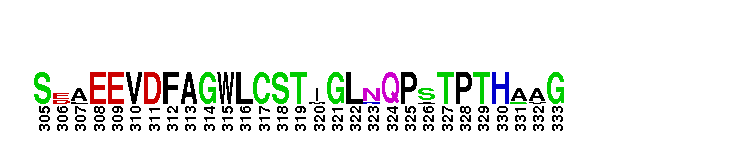| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tips:  Range on the Protein: Protein ID Protein Position Domain Position: 
|
|---|
Weblogos are Copyright (c) 2002 Regents of the University of California
| DMDM_info@umbc.edu | 1000 Hilltop Circle, Baltimore, MD 21250 | Department of Biological Sciences | Phone: 410-455-2258 |




 Catalytic domain of the dual-specificity Protein Kinase, MAP/ERK Kinase 1. Protein kinases (PKs), MAP/ERK kinase (MEK) 1 subfamily, catalytic (c) domain. PKs catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine or tyrosine residues on protein substrates. The MEK subfamily is part of a larger superfamily that includes the catalytic domains of other protein serine/threonine kinases, protein tyrosine kinases, RIO kinases, aminoglycoside phosphotransferase, choline kinase, and phosphoinositide 3-kinase. The mitogen-activated protein (MAP) kinase signaling pathways are important mediators of cellular responses to extracellular signals. The pathways involve a triple kinase core cascade comprising the MAP kinase (MAPK), which is phosphorylated and activated by a MAPK kinase (MAPKK or MKK), which itself is phosphorylated and activated by a MAPK kinase kinase (MAPKKK or MKKK). MEK1 is a dual-specificity PK that phosphorylates and activates the downstream targets, extracellular signal-regulated kinase (ERK) 1 and ERK2, on specific threonine and tyrosine residues. The ERK cascade starts with extracellular signals including growth factors, hormones, and neurotransmitters, which act through receptors and ion channels to initiate intracellular signaling that leads to the activation at the MAPKKK (Raf-1 or MOS) level, which leads to the transmission of signals to MEK1, and finally to ERK1/2. The ERK cascade plays an important role in cell proliferation, differentiation, oncogenic transformation, and cell cycle control, as well as in apoptosis and cell survival under certain conditions. Gain-of-function mutations in genes encoding ERK cascade proteins, including MEK1, cause cardiofaciocutaneous (CFC) syndrome, a condition leading to multiple congenital anomalies and mental retardation in patients. MEK1 also plays a role in cell cycle control.
Catalytic domain of the dual-specificity Protein Kinase, MAP/ERK Kinase 1. Protein kinases (PKs), MAP/ERK kinase (MEK) 1 subfamily, catalytic (c) domain. PKs catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine or tyrosine residues on protein substrates. The MEK subfamily is part of a larger superfamily that includes the catalytic domains of other protein serine/threonine kinases, protein tyrosine kinases, RIO kinases, aminoglycoside phosphotransferase, choline kinase, and phosphoinositide 3-kinase. The mitogen-activated protein (MAP) kinase signaling pathways are important mediators of cellular responses to extracellular signals. The pathways involve a triple kinase core cascade comprising the MAP kinase (MAPK), which is phosphorylated and activated by a MAPK kinase (MAPKK or MKK), which itself is phosphorylated and activated by a MAPK kinase kinase (MAPKKK or MKKK). MEK1 is a dual-specificity PK that phosphorylates and activates the downstream targets, extracellular signal-regulated kinase (ERK) 1 and ERK2, on specific threonine and tyrosine residues. The ERK cascade starts with extracellular signals including growth factors, hormones, and neurotransmitters, which act through receptors and ion channels to initiate intracellular signaling that leads to the activation at the MAPKKK (Raf-1 or MOS) level, which leads to the transmission of signals to MEK1, and finally to ERK1/2. The ERK cascade plays an important role in cell proliferation, differentiation, oncogenic transformation, and cell cycle control, as well as in apoptosis and cell survival under certain conditions. Gain-of-function mutations in genes encoding ERK cascade proteins, including MEK1, cause cardiofaciocutaneous (CFC) syndrome, a condition leading to multiple congenital anomalies and mental retardation in patients. MEK1 also plays a role in cell cycle control. No pairwise interactions are available for this conserved domain.
No pairwise interactions are available for this conserved domain.