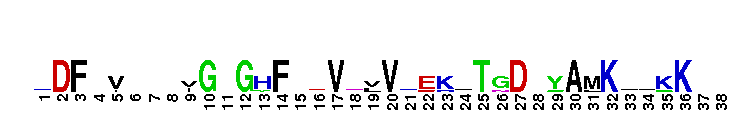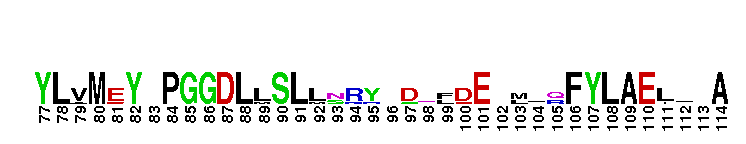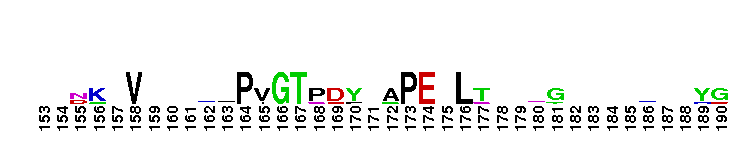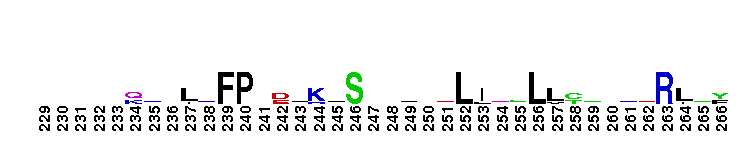AMYOTROPHIC LATERAL SCLEROSIS 19 AMYOTROPHIC LATERAL SCLEROSIS 19
|
 AORTIC ANEURYSM, FAMILIAL THORACIC 7 AORTIC ANEURYSM, FAMILIAL THORACIC 7
|
 BREAST CANCER, SUSCEPTIBILITY TO BREAST CANCER, SUSCEPTIBILITY TO
|
 CARDIOFACIOCUTANEOUS SYNDROME 3 CARDIOFACIOCUTANEOUS SYNDROME 3
|
 CARDIOFACIOCUTANEOUS SYNDROME 4 CARDIOFACIOCUTANEOUS SYNDROME 4
|
 CATARACT 6, AGE-RELATED CORTICAL CATARACT 6, AGE-RELATED CORTICAL
|
 CEREBRAL INFARCTION, SUSCEPTIBILITY TO CEREBRAL INFARCTION, SUSCEPTIBILITY TO
|
 CHRONIC MYELOID LEUKEMIA, RESISTANT TO IMATINIB CHRONIC MYELOID LEUKEMIA, RESISTANT TO IMATINIB
|
 COFFIN-LOWRY SYNDROME COFFIN-LOWRY SYNDROME
|
 COFFIN-LOWRY SYNDROME, MILD COFFIN-LOWRY SYNDROME, MILD
|
 COLON CANCER, HEREDITARY NONPOLYPOSIS, TYPE 6, SOMATIC, INCLUDED COLON CANCER, HEREDITARY NONPOLYPOSIS, TYPE 6, SOMATIC, INCLUDED
|
 COLORECTAL CANCER, HEREDITARY NONPOLYPOSIS, TYPE 6 COLORECTAL CANCER, HEREDITARY NONPOLYPOSIS, TYPE 6
|
 COWDEN DISEASE 6 COWDEN DISEASE 6
|
 DIABETES MELLITUS, INSULIN-RESISTANT, WITH ACANTHOSIS NIGRICANS DIABETES MELLITUS, INSULIN-RESISTANT, WITH ACANTHOSIS NIGRICANS
|
 DIABETES MELLITUS, NONINSULIN-DEPENDENT DIABETES MELLITUS, NONINSULIN-DEPENDENT
|
 DIABETES MELLITUS, TYPE II DIABETES MELLITUS, TYPE II
|
 ENDOCRINE-CEREBROOSTEODYSPLASIA ENDOCRINE-CEREBROOSTEODYSPLASIA
|
 EPILEPTIC ENCEPHALOPATHY, EARLY INFANTILE, 2 EPILEPTIC ENCEPHALOPATHY, EARLY INFANTILE, 2
|
 ESOPHAGEAL CANCER, SOMATIC ESOPHAGEAL CANCER, SOMATIC
|
 FG SYNDROME 4 FG SYNDROME 4
|
 GLYCOGEN STORAGE DISEASE IXC GLYCOGEN STORAGE DISEASE IXC
|
 HYPERINSULINEMIC HYPOGLYCEMIA, FAMILIAL, 5 HYPERINSULINEMIC HYPOGLYCEMIA, FAMILIAL, 5
|
 INSULIN RESISTANCE INSULIN RESISTANCE
|
 INSULIN RESISTANCE, INCLUDED INSULIN RESISTANCE, INCLUDED
|
 IRAK4 DEFICIENCY IRAK4 DEFICIENCY
|
 LEUKEMIA, PHILADELPHIA CHROMOSOME-POSITIVE, RESISTANT TO IMATINIB LEUKEMIA, PHILADELPHIA CHROMOSOME-POSITIVE, RESISTANT TO IMATINIB
|
 LOEYS-DIETZ SYNDROME, TYPE 1A LOEYS-DIETZ SYNDROME, TYPE 1A
|
 LOEYS-DIETZ SYNDROME, TYPE 1B LOEYS-DIETZ SYNDROME, TYPE 1B
|
 LOEYS-DIETZ SYNDROME, TYPE 1B, INCLUDED LOEYS-DIETZ SYNDROME, TYPE 1B, INCLUDED
|
 LOEYS-DIETZ SYNDROME, TYPE 2A LOEYS-DIETZ SYNDROME, TYPE 2A
|
 LOEYS-DIETZ SYNDROME, TYPE 2A, INCLUDED LOEYS-DIETZ SYNDROME, TYPE 2A, INCLUDED
|
 LOEYS-DIETZ SYNDROME, TYPE 2B LOEYS-DIETZ SYNDROME, TYPE 2B
|
 MALFORMATIONS MALFORMATIONS
|
 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME
|
 MELANOMA, MALIGNANT, SOMATIC MELANOMA, MALIGNANT, SOMATIC
|
 MENTAL RETARDATION AND MICROCEPHALY WITH PONTINE AND CEREBELLAR HYPOPLASIA MENTAL RETARDATION AND MICROCEPHALY WITH PONTINE AND CEREBELLAR HYPOPLASIA
|
 MENTAL RETARDATION, X-LINKED 19 MENTAL RETARDATION, X-LINKED 19
|
 MENTAL RETARDATION, X-LINKED 30 MENTAL RETARDATION, X-LINKED 30
|
 MENTAL RETARDATION, X-LINKED, WITH NYSTAGMUS MENTAL RETARDATION, X-LINKED, WITH NYSTAGMUS
|
 MULTIPLE SELF-HEALING SQUAMOUS EPITHELIOMA, SUSCEPTIBILITY TO MULTIPLE SELF-HEALING SQUAMOUS EPITHELIOMA, SUSCEPTIBILITY TO
|
 NEPHRONOPHTHISIS 9 (NPHP9) NEPHRONOPHTHISIS 9 (NPHP9)
|
 NEUROPATHY, HEREDITARY SENSORY, TYPE II NEUROPATHY, HEREDITARY SENSORY, TYPE II
|
 OGUCHI DISEASE 2 OGUCHI DISEASE 2
|
 PEUTZ-JEGHERS SYNDROME PEUTZ-JEGHERS SYNDROME
|
 PROSTATE CANCER, PROGRESSION AND METASTASIS OF PROSTATE CANCER, PROGRESSION AND METASTASIS OF
|
 RETINITIS PIGMENTOSA 62 RETINITIS PIGMENTOSA 62
|
 RETINITIS PIGMENTOSA 62 (RP62) RETINITIS PIGMENTOSA 62 (RP62)
|
 SELECTIVE T-CELL DEFECT SELECTIVE T-CELL DEFECT
|
 SHORT RIB-POLYDACTYLY SYNDROME, TYPE IIA SHORT RIB-POLYDACTYLY SYNDROME, TYPE IIA
|
 SPERMATOGENIC FAILURE 5 SPERMATOGENIC FAILURE 5
|
 SPINOCEREBELLAR ATAXIA 14 SPINOCEREBELLAR ATAXIA 14
|
 T-CELL IMMUNODEFICIENCY, RECURRENT INFECTIONS, AUTOIMMUNITY, AND CARDIAC T-CELL IMMUNODEFICIENCY, RECURRENT INFECTIONS, AUTOIMMUNITY, AND CARDIAC
|
 TESTICULAR TUMOR, SOMATIC TESTICULAR TUMOR, SOMATIC
|
 THROMBOCYTOPENIA 2 THROMBOCYTOPENIA 2
|
 VARIANT OF UNKNOWN SIGNIFICANCE VARIANT OF UNKNOWN SIGNIFICANCE
|
 VENOUS MALFORMATIONS, MULTIPLE CUTANEOUS AND MUCOSAL VENOUS MALFORMATIONS, MULTIPLE CUTANEOUS AND MUCOSAL
|






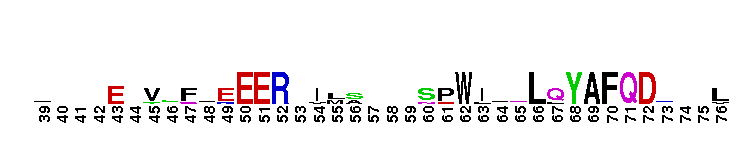
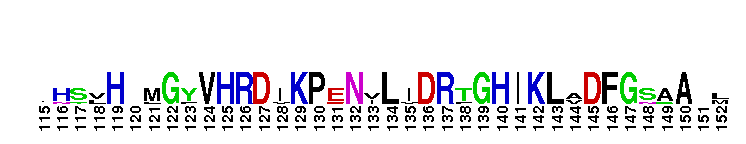
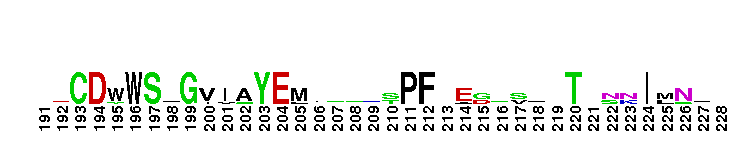
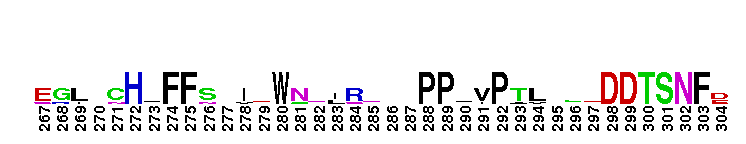
 Catalytic domain of the Protein Serine/Threonine Kinase, Citron Rho-interacting kinase. Serine/Threonine Kinases (STKs), Citron Rho-interacting kinase (CRIK) subfamily, catalytic (c) domain. STKs catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine residues on protein substrates. The CRIK subfamily is part of a larger superfamily that includes the catalytic domains of other protein STKs, protein tyrosine kinases, RIO kinases, aminoglycoside phosphotransferase, choline kinase, and phosphoinositide 3-kinase. CRIK is also called citron kinase. It contains a catalytic domain, a central coiled-coil domain, and a C-terminal region containing a Rho-binding domain (RBD), a zinc finger, and a pleckstrin homology (PH) domain, in addition to other motifs. CRIK, an effector of the small GTPase Rho, plays an important function during cytokinesis and affects its contractile process. CRIK-deficient mice show severe ataxia and epilepsy as a result of abnormal cytokinesis and massive apoptosis in neuronal precursors. A Down syndrome critical region protein TTC3 interacts with CRIK and inhibits CRIK-dependent neuronal differentiation and neurite extension.
Catalytic domain of the Protein Serine/Threonine Kinase, Citron Rho-interacting kinase. Serine/Threonine Kinases (STKs), Citron Rho-interacting kinase (CRIK) subfamily, catalytic (c) domain. STKs catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine residues on protein substrates. The CRIK subfamily is part of a larger superfamily that includes the catalytic domains of other protein STKs, protein tyrosine kinases, RIO kinases, aminoglycoside phosphotransferase, choline kinase, and phosphoinositide 3-kinase. CRIK is also called citron kinase. It contains a catalytic domain, a central coiled-coil domain, and a C-terminal region containing a Rho-binding domain (RBD), a zinc finger, and a pleckstrin homology (PH) domain, in addition to other motifs. CRIK, an effector of the small GTPase Rho, plays an important function during cytokinesis and affects its contractile process. CRIK-deficient mice show severe ataxia and epilepsy as a result of abnormal cytokinesis and massive apoptosis in neuronal precursors. A Down syndrome critical region protein TTC3 interacts with CRIK and inhibits CRIK-dependent neuronal differentiation and neurite extension. No pairwise interactions are available for this conserved domain.
No pairwise interactions are available for this conserved domain.